Ishigami Function
Contents
Ishigami Function#
import numpy as np
import matplotlib.pyplot as plt
import uqtestfuns as uqtf
The Ishigami test function is a three-dimensional scalar-valued function. First introduced in [IH91] in the context of sensitivity analysis, the function has been revisited many times in the same context (see for instances [SobolL99, Sud08, MILR09]).
Test function instance#
To create a default instance of the Ishigami test function:
my_testfun = uqtf.Ishigami()
Check if it has been correctly instantiated:
print(my_testfun)
Name : Ishigami
Spatial dimension : 3
Description : Ishigami function from Ishigami and Homma (1991)
Description#
The Ishigami function, a highly non-linear and non-monotonic function, is given as follows:
where \(\boldsymbol{x} = \{ x_1, x_2, x_3 \}\) is the three-dimensional vector of input variables further defined below, and \(a\) and \(b\) are parameters of the function.
Probabilistic input#
Based on [IH91], the probabilistic input model for the Ishigami function consists of three independent uniform random variables with the ranges shown in the table below.
my_testfun.prob_input
Name: Ishigami1991
Spatial Dimension: 3
Description: Probabilistic input model for the Ishigami function from Ishigami and Homma (1991).
Marginals:
| No. | Name | Distribution | Parameters | Description |
|---|---|---|---|---|
| 1 | X1 | uniform | [-3.14159265 3.14159265] | None |
| 2 | X2 | uniform | [-3.14159265 3.14159265] | None |
| 3 | X3 | uniform | [-3.14159265 3.14159265] | None |
Copulas: None
Parameters#
The parameters of the Ishigami function are two real-valued numbers. Some of the available parameter values taken from the literature are shown in the table below.
No. |
Value |
Keyword |
Source |
|---|---|---|---|
1 |
\(a = 7\), \(b = 0.1\) |
|
[IH91] |
2 |
\(a = 7\), \(b = 0.05\) |
|
[SobolL99] |
Alternatively, to create an instance of the Ishigami function with different parameter values, type:
my_testfun = uqtf.Ishigami(parameters_selection="Sobol1999")
Note
To use another set of parameters, create a default test function
and pass one of the available keywords
(as indicated in the table above) to the parameters_selection parameter.
For example:
my_testfun = uqtf.Ishigami(parameters_selection="Sobol1999")
Reference results#
This section provides several reference results of typical UQ analyses involving the test function.
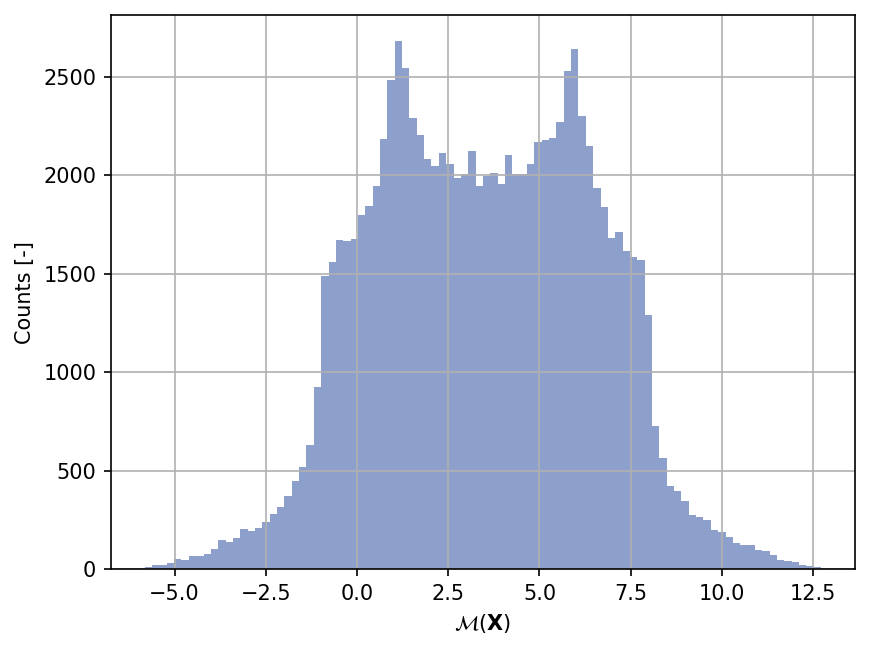
Sample histogram#
Shown below is the histogram of the output based on \(100'000\) random points:
np.random.seed(42)
xx_test = my_testfun.prob_input.get_sample(100000)
yy_test = my_testfun(xx_test)
plt.hist(yy_test, bins="auto", color="#8da0cb");
plt.grid();
plt.ylabel("Counts [-]");
plt.xlabel("$\mathcal{M}(\mathbf{X})$");
plt.gcf().set_dpi(150);
Moment estimations#
The mean and variance of the Ishigami function can be computed analytically,
and the results are:
\(\mathbb{E}[Y] = \frac{a}{2}\)
\(\mathbb{V}[Y] = \frac{a^2}{8} + \frac{b \pi^4}{5} + \frac{b^2 \pi^8}{18} + \frac{1}{2}\)
Notice that the values of these two moments depend on the choice of the parameter values.
Shown below is the convergence of a direct Monte-Carlo estimation of the output mean and variance with increasing sample sizes compared with the analytical values.
# --- Compute the mean and variance estimate
np.random.seed(42)
sample_sizes = np.array([1e1, 1e2, 1e3, 1e4, 1e5, 1e6, 1e7], dtype=int)
mean_estimates = np.empty(len(sample_sizes))
var_estimates = np.empty(len(sample_sizes))
for i, sample_size in enumerate(sample_sizes):
xx_test = my_testfun.prob_input.get_sample(sample_size)
yy_test = my_testfun(xx_test)
mean_estimates[i] = np.mean(yy_test)
var_estimates[i] = np.var(yy_test)
# --- Compute the error associated with the estimates
mean_estimates_errors = np.sqrt(var_estimates) / np.sqrt(np.array(sample_sizes))
var_estimates_errors = var_estimates * np.sqrt(2 / (np.array(sample_sizes) - 1))
fig, ax_1 = plt.subplots(figsize=(6,4))
# --- Mean plot
ax_1.errorbar(
sample_sizes,
mean_estimates,
yerr=mean_estimates_errors,
marker="o",
color="#66c2a5",
label="Mean"
)
# Plot the analytical mean
a = my_testfun.parameters[0]
mean_analytical = a / 2.0
ax_1.plot(
sample_sizes,
np.repeat(mean_analytical, len(sample_sizes)),
linestyle="--",
color="#66c2a5",
label="Analytical mean",
)
ax_1.set_xlim([9, 2e7])
ax_1.set_xlabel("Sample size")
ax_1.set_ylabel("Output mean estimate")
ax_1.set_xscale("log");
ax_2 = ax_1.twinx()
# --- Variance plot
ax_2.errorbar(
sample_sizes+1,
var_estimates,
yerr=var_estimates_errors,
marker="o",
color="#fc8d62",
label="Variance",
)
# Plot the analytical variance
b = my_testfun.parameters[1]
var_analytical = a**2 / 8 + b * np.pi**4 / 5 + b**2 * np.pi**8 / 18 + 0.5
ax_2.plot(
sample_sizes,
np.repeat(var_analytical, len(sample_sizes)),
linestyle="--",
color="#fc8d62",
label="Analytical variance",
)
ax_2.set_ylabel("Output variance estimate")
# Add the two plots together to have a common legend
ln_1, labels_1 = ax_1.get_legend_handles_labels()
ln_2, labels_2 = ax_2.get_legend_handles_labels()
ax_2.legend(ln_1 + ln_2, labels_1 + labels_2, loc=0)
plt.grid()
fig.set_dpi(150)
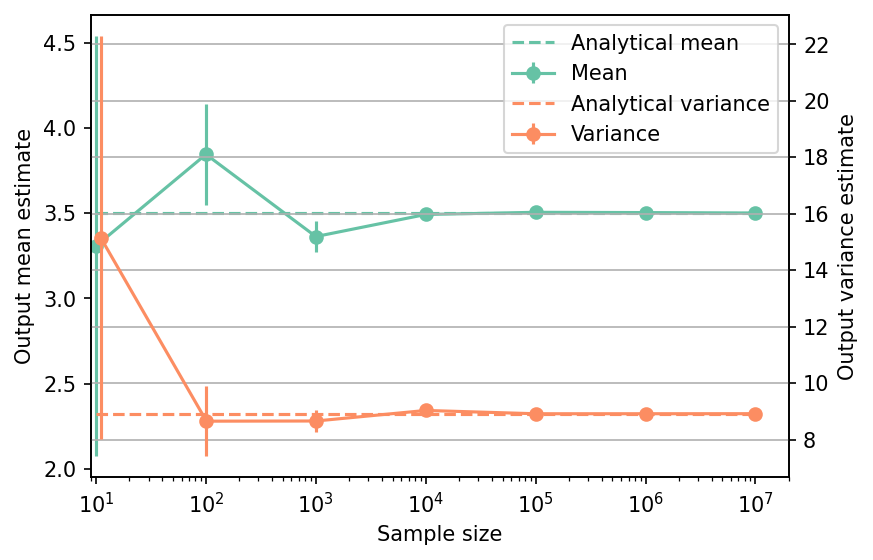
The tabulated results for each sample size is shown below.
from tabulate import tabulate
# --- Compile data row-wise
outputs = [
[
np.nan,
mean_analytical,
0.0,
var_analytical,
0.0,
"Analytical",
]
]
for (
sample_size,
mean_estimate,
mean_estimate_error,
var_estimate,
var_estimate_error,
) in zip(
sample_sizes,
mean_estimates,
mean_estimates_errors,
var_estimates,
var_estimates_errors,
):
outputs += [
[
sample_size,
mean_estimate,
mean_estimate_error,
var_estimate,
var_estimate_error,
"Monte-Carlo",
],
]
header_names = [
"Sample size",
"Mean",
"Mean error",
"Variance",
"Variance error",
"Remark",
]
tabulate(
outputs,
headers=header_names,
floatfmt=(".1e", ".4e", ".4e", ".4e", ".4e", "s"),
tablefmt="html",
stralign="center",
numalign="center",
)
| Sample size | Mean | Mean error | Variance | Variance error | Remark |
|---|---|---|---|---|---|
| nan | 3.5000e+00 | 0.0000e+00 | 8.9169e+00 | 0.0000e+00 | Analytical |
| 1.0e+01 | 3.3079e+00 | 1.2312e+00 | 1.5159e+01 | 7.1458e+00 | Monte-Carlo |
| 1.0e+02 | 3.8445e+00 | 2.9416e-01 | 8.6529e+00 | 1.2299e+00 | Monte-Carlo |
| 1.0e+03 | 3.3626e+00 | 9.3054e-02 | 8.6591e+00 | 3.8744e-01 | Monte-Carlo |
| 1.0e+04 | 3.4921e+00 | 3.0054e-02 | 9.0327e+00 | 1.2775e-01 | Monte-Carlo |
| 1.0e+05 | 3.5048e+00 | 9.4424e-03 | 8.9160e+00 | 3.9874e-02 | Monte-Carlo |
| 1.0e+06 | 3.5032e+00 | 2.9860e-03 | 8.9164e+00 | 1.2610e-02 | Monte-Carlo |
| 1.0e+07 | 3.5007e+00 | 9.4437e-04 | 8.9184e+00 | 3.9884e-03 | Monte-Carlo |
Sensitivity indices#
The main-effect and total-effect Sobol’ indices of the Ishigami function can be
derived analytically.
The main-effect (i.e., first-order) Sobol’ indices are:
\(S_1 \equiv \frac{V_1}{\mathbb{V}[Y]}\)
\(S_2 \equiv \frac{V_2}{\mathbb{V}[Y]}\)
\(S_3 \equiv \frac{V_3}{\mathbb{V}[Y]}\)
where the total variances \(\mathbb{V}[Y]\) is given in the section above and
the partial variances are given by:
\(V_1 = \frac{1}{2} \, (1 + \frac{b \pi^4}{5})^2\)
\(V_2 = \frac{a^2}{8}\)
\(V_3 = 0\)
The total-effect Sobol’ indices, on the other hand:
\(ST_1 \equiv \frac{VT_1}{\mathbb{V}[Y]}\)
\(ST_2 \equiv \frac{VT_2}{\mathbb{V}[Y]}\)
\(ST_3 \equiv \frac{VT_3}{\mathbb{V}[Y]}\)
where:
\(VT_1 = \frac{1}{2} \, (1 + \frac{b \pi^4}{5})^2 + \frac{8 b^2 \pi^8}{225}\)
\(VT_2 = \frac{a^2}{8}\)
\(VT_3 = \frac{8 b^2 \pi^8}{225}\)
Note, once more, that the values of the partial variances depend on the parameter values.
Note
The Ishigami function has a peculiar dependence on \(X_3\); the main-effect index of the input variable is \(0\) but the total-effect index is not, due to an interaction with \(X_1\)!
Furthermore, as can be seen from the function definition, \(X_2\) has no interaction effect; its main-effect and total-effect indices are exactly the same.
References#
- IH91(1,2,3)
T. Ishigami and T. Homma. An importance quantification technique in uncertainty analysis for computer models. In [1990] Proceedings. First International Symposium on Uncertainty Modeling and Analysis, 398–403. IEEE Comput. Soc. Press, 1991. doi:10.1109/ISUMA.1990.151285.
- SobolL99(1,2)
Ilya M. Sobol' and Yu L. Levitan. On the use of variance reducing multipliers in Monte Carlo computations of a global sensitivity index. Computer Physics Communications, 117(1):52–61, 1999. doi:10.1016/S0010-4655(98)00156-8.
- Sud08
Bruno Sudret. Global sensitivity analysis using polynomial chaos expansions. Reliability Engineering & System Safety, 93(7):964–979, 2008. doi:10.1016/j.ress.2007.04.002.
- MILR09
Amandine Marrel, Bertrand Iooss, Béatrice Laurent, and Olivier Roustant. Calculations of Sobol indices for the Gaussian process metamodel. Reliability Engineering & System Safety, 94(3):742–751, 2009. doi:10.1016/j.ress.2008.07.008.

