Oakley and O’Hagan (2002) One-dimensional (1D) Function
Contents
Oakley and O’Hagan (2002) One-dimensional (1D) Function#
The 1D function from Oakley and O’Hagan (2002) (or Oakley1D function
for short) is a scalar-valued test function.
It was used in [OOHagan02] as a test function for illustrating metamodeling
and uncertainty propagation approaches.
import numpy as np
import matplotlib.pyplot as plt
import uqtestfuns as uqtf
A plot of the function is shown below for \(x \in [-12, 12]\).
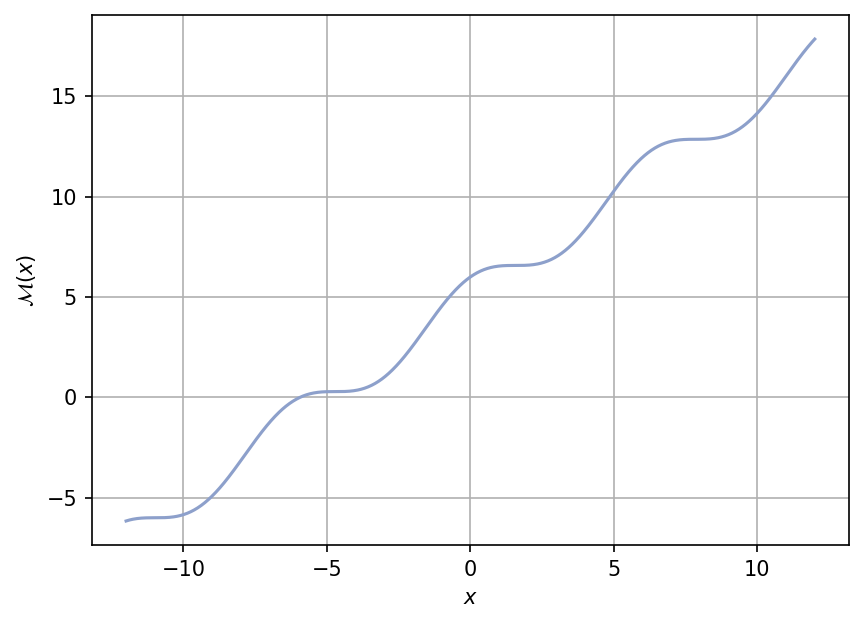
Test function instance#
To create a default instance of the test function:
my_testfun = uqtf.Oakley1D()
Check if it has been correctly instantiated:
print(my_testfun)
Name : Oakley1D
Spatial dimension : 1
Description : One-dimensional function from Oakley and O'Hagan (2002)
Description#
The test function is analytically defined as follows:
where \(x\) is probabilistically defined below.
Probabilistic input#
Based on [OOHagan02], the probabilistic input model for the 1D Oakley-O’Hagan function consists of a normal random variable with the parameters shown in the table below.
my_testfun.prob_input
Name: Oakley2002
Spatial Dimension: 1
Description: Probabilistic input model for the one-dimensional function from Oakley and O'Hagan (2002)
Marginals:
| No. | Name | Distribution | Parameters | Description |
|---|---|---|---|---|
| 1 | x | normal | [0. 4.] | None |
Copulas: None
Reference results#
This section provides several reference results of typical UQ analyses involving the test function.
Sample histogram#
Shown below is the histogram of the output based on \(100'000\) random points:
np.random.seed(42)
xx_test = my_testfun.prob_input.get_sample(100000)
yy_test = my_testfun(xx_test)
plt.hist(yy_test, bins="auto", color="#8da0cb");
plt.grid();
plt.ylabel("Counts [-]");
plt.xlabel("$\mathcal{M}(X)$");
plt.gcf().tight_layout(pad=3.0)
plt.gcf().set_dpi(150);
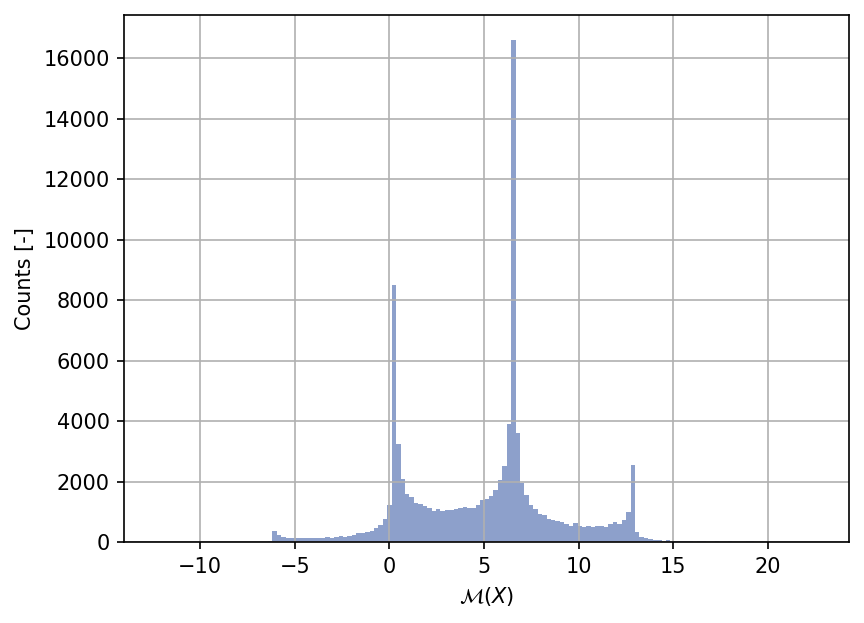
Moment estimations#
Shown below is the convergence of a direct Monte-Carlo estimation of the output mean and variance with increasing sample sizes.
np.random.seed(42)
sample_sizes = np.array([1e1, 1e2, 1e3, 1e4, 1e5, 1e6], dtype=int)
mean_estimates = np.empty((len(sample_sizes), 50))
var_estimates = np.empty((len(sample_sizes), 50))
for i, sample_size in enumerate(sample_sizes):
for j in range(50):
xx_test = my_testfun.prob_input.get_sample(sample_size)
yy_test = my_testfun(xx_test)
mean_estimates[i, j] = np.mean(yy_test)
var_estimates[i, j] = np.var(yy_test)
# --- Compute the error associated with the estimates
mean_estimates_errors = np.std(mean_estimates, axis=1)
var_estimates_errors = np.std(var_estimates, axis=1)
# --- Plot the mean and variance estimates
fig, ax_1 = plt.subplots(figsize=(6,4))
# --- Mean plot
ax_1.errorbar(
sample_sizes,
mean_estimates[:,0],
yerr=2.0*mean_estimates_errors,
marker="o",
color="#66c2a5",
label="Mean"
)
ax_1.set_xlim([5, 2e6])
ax_1.set_xlabel("Sample size")
ax_1.set_ylabel("Output mean estimate")
ax_1.set_xscale("log");
ax_2 = ax_1.twinx()
# --- Variance plot
ax_2.errorbar(
sample_sizes+1,
var_estimates[:,0],
yerr=1.96*var_estimates_errors,
marker="o",
color="#fc8d62",
label="Variance",
)
ax_2.set_ylabel("Output variance estimate")
# Add the two plots together to have a common legend
ln_1, labels_1 = ax_1.get_legend_handles_labels()
ln_2, labels_2 = ax_2.get_legend_handles_labels()
ax_2.legend(ln_1 + ln_2, labels_1 + labels_2, loc=0)
plt.grid()
fig.set_dpi(150)
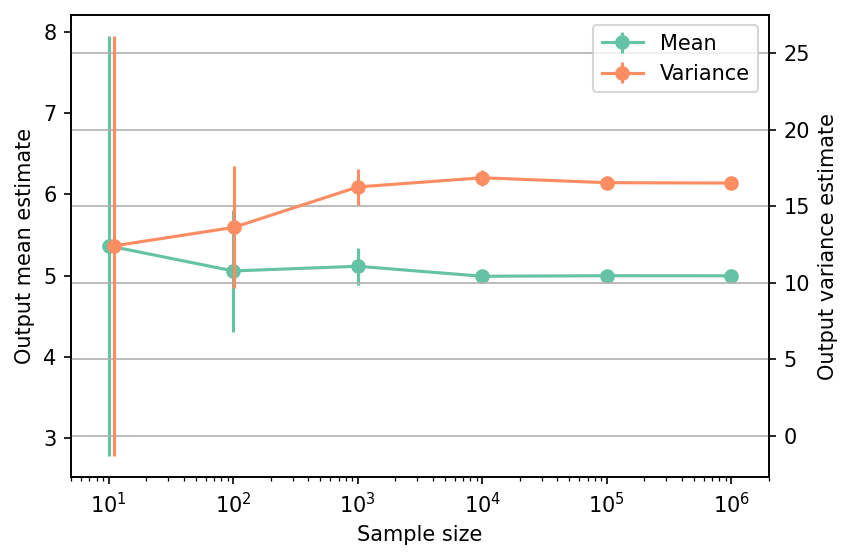
The tabulated results for each sample size is shown below.
from tabulate import tabulate
# --- Compile data row-wise
outputs =[]
for (
sample_size,
mean_estimate,
mean_estimate_error,
var_estimate,
var_estimate_error,
) in zip(
sample_sizes,
mean_estimates[:,0],
2.0*mean_estimates_errors,
var_estimates[:,0],
2.0*var_estimates_errors,
):
outputs += [
[
sample_size,
mean_estimate,
mean_estimate_error,
var_estimate,
var_estimate_error,
"Monte-Carlo",
],
]
header_names = [
"Sample size",
"Mean",
"Mean error",
"Variance",
"Variance error",
"Remark",
]
tabulate(
outputs,
numalign="center",
stralign="center",
tablefmt="html",
floatfmt=(".1e", ".4e", ".4e", ".4e", ".4e", "s"),
headers=header_names
)
| Sample size | Mean | Mean error | Variance | Variance error | Remark |
|---|---|---|---|---|---|
| 1.0e+01 | 5.3667e+00 | 2.5821e+00 | 1.2412e+01 | 1.4007e+01 | Monte-Carlo |
| 1.0e+02 | 5.0586e+00 | 7.4662e-01 | 1.3622e+01 | 4.0659e+00 | Monte-Carlo |
| 1.0e+03 | 5.1161e+00 | 2.2424e-01 | 1.6265e+01 | 1.1872e+00 | Monte-Carlo |
| 1.0e+04 | 4.9933e+00 | 8.0880e-02 | 1.6864e+01 | 5.4213e-01 | Monte-Carlo |
| 1.0e+05 | 4.9998e+00 | 2.5349e-02 | 1.6542e+01 | 1.3712e-01 | Monte-Carlo |
| 1.0e+06 | 4.9988e+00 | 7.3062e-03 | 1.6518e+01 | 4.4011e-02 | Monte-Carlo |
References#
- OOHagan02(1,2)
Jeremy Oakley and Anthony O'Hagan. Bayesian inference for the uncertainty distribution of computer model outputs. Biometrika, 89(4):769–784, 2002. doi:10.1093/biomet/89.4.769.

